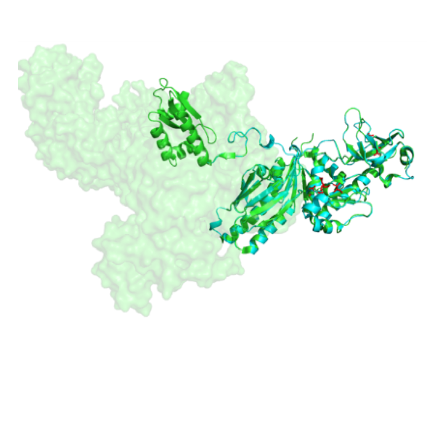
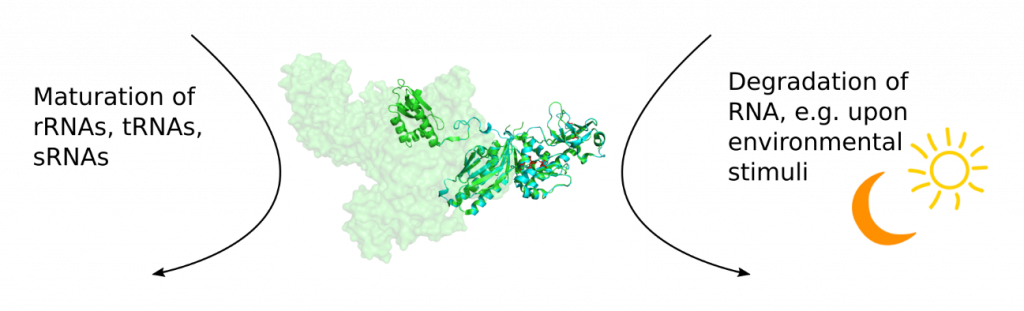
Crystal structure of E. coli RNase E (green, pdb 2vmk) and a homology model of Synechocystis RNase E (blue, using SWISS Model).
RNA maturation and degradation are key aspects of RNA metabolism. Endoribonuclease RNase E is involved in both processes. In E. coli, where it was mainly investigated, RNase E is essential for tRNA, rRNA and sRNA maturation. Also, it is scaffolding the membrane-bound degradosome and is an integral enzyme in RNA degradation.
The role of Synechocystis RNase E is, so far, less understood than the ones of E. coli RNase E. Synechocystis RNase E shares functions with E. coli RNase E, such as being involved in tRNA and rRNA maturation and the action of some sRNAs. However, some facets differ strikingly. Examples are its cellular localization and its involvement in the maturation of the CRISPR3 system. Adding to this, the protein Hfq, which is an RNA chaperone mediating the interaction of RNase E and many sRNAs in E. coli, fulfills a completely different function in Synechocystis.
We investigate the target affinity of RNase E in Synechocystis by using high-throughput RNA sequencing techniques (e.g. iCLIP, TIER-Seq) and in vitro cleavage assays. Furthermore, we perform phenotypic studies of mutant strains and fluorescence microscopy to elucidate its cellular localization.
Funding:
- GRK2344 „MeInBio – Exploration of spatio-temporal dynamics of gene regulation using high-throughput and high-resolution methods”
Key publications:
Hoffmann UA, Heyl F, Rogh SN, Wallner T, Backofen R, Hess WR, Steglich C, Wilde A. (2021) Transcriptome-wide in vivo mapping of cleavage sites for the compact cyanobacterial ribonuclease E reveals insights into its function and substrate recognition. Nucleic Acids Res. 49, 13075-13091. doi: 10.1093/nar/gkab1161.
Mahbub M, Hemm L, Yang Y, Kaur R, Carmen H, Engl C, Huokko T, Riediger M, Watanabe S, Liu LN, Wilde A, Hess WR, Mullineaux CW. (2020) mRNA localization, reaction centre biogenesis and thylakoid membrane targeting in cyanobacteria. Nat Plants. 6:1179-1191.
Behler J, Sharma K, Reimann V, Wilde A, Urlaub H, Hess WR. (2018) The host-encoded RNase E endonuclease as the crRNA maturation enzyme in a CRISPR-Cas subtype III-Bv system. Nat. Microbiol. 3, 367-377
Wilde A, Hihara Y. Transcriptional and posttranscriptional regulation of cyanobacterial photosynthesis. (2015) Biochim Biophys Acta. doi: 10.1016/j.bbabio.2015.11.002. [Epub ahead of print]
Georg J, Dienst D, Schuergers N, Wallner T, Kopp D, Stazic D, Kuchmina K, Klähn S, Lokstein H, Hess WR, Wilde A. (2014) The small regulatory RNA SyR1/PsrR1 controls photosynthetic functions in cyanobacteria. Plant Cell 26, 3661-79.
Hess WR, Berghoff BA, Wilde A, Steglich C, Klug G. (2014) Riboregulators and the role of Hfq in photosynthetic bacteria. RNA Biol. 11, 413-425.
Schuergers N, Ruppert U, Watanabe S, Nürnberg DJ, Lochnit G, Dienst D, Mullineaux CW, Wilde A. (2014) Binding of the RNA chaperone Hfq to the type IV pilus base is crucial for its function in Synechocystis sp. PCC 6803. Mol. Microbiol. 92, 840-852.
Mitschke, J., Georg, J., Scholz, I., Sharma, C.M., Dienst, D., Bantscheff, J., Voß, B., Steglich, C., Wilde, A., Vogel, J., Hess, W.R. (2011) An experimentally anchored map of transcriptional start sites in the model cyanobacterium Synechocystis sp. PCC 6803. Proc Natl Acad Sci U S A. 108, 2124-2129.
Georg, J., Voss, B., Scholz, I., Mitschke, J., Wilde, A., Hess, W.R. (2009) Evidence for a major role of antisense RNAs in cyanobacterial gene regulation. Molecular Systems Biology 5, 305.
Legewie, S., Dienst, D., Wilde, A., Herzel, H., Axmann, I.M. (2008) Small RNAs establish delays and temporal thresholds in gene expression. Biophys J. 95, 3232-3238.
Dienst, D., Dühring, U., Mollenkopf, H.J., Vogel, J., Golecki, J., Hess, W.R., Wilde, A. (2008) The cyanobacterial homologue of the RNA chaperone Hfq is essential for motility of Synechocystis sp. PCC 6803. Microbiology 154, 3134-3143
Dühring, U., Axmann, I.M., Hess, W.R., Wilde, A. (2006) A naturally occurring antisense RNA affecting expression of isiA and assembly of IsiA supercomplexes. Proc. Nat. Acad. Sci. 103, 7054-7058.
Wilde, A. (2008) Antisense-RNAs in Bakterien. Naturwissenschaftliche Rundschau 61, 226-229