The coordination of biological activities into daily cycles provides an important advantage for the fitness of diverse organisms. An internal circadian oscillator drives gene expression in an approximate 24 hours rhythm. For the cyanobacterium Synechococcus elongatus PCC 7942, only three different cyanobacterial proteins (KaiA, KaiB, and KaiC) are sufficient to achieve a circadian rhythm of phosphorylation cycles in vitro. KaiC as the major clock component has three intrinsic enzymatic activities: autokinase, autophosphatase and ATPase activities, whereas KaiA enhances the kinase and KaiB the autophosphatase activities, respectively. Most data on the circadian timing process in cyanobacteria have been obtained using the unicellular cyanobacterium Synechococcus elongatus. Other model cyanobacteria like Synechocystis sp. PCC 6803 harbor multiple gene copies for the three clock components. In this project, we analyze the main oscillator KaiAB1C1, focusing on the signaling-output pathway and the non-standard circadian clocks KaiB2C2 and KaiB3C3 of Synechocystis.
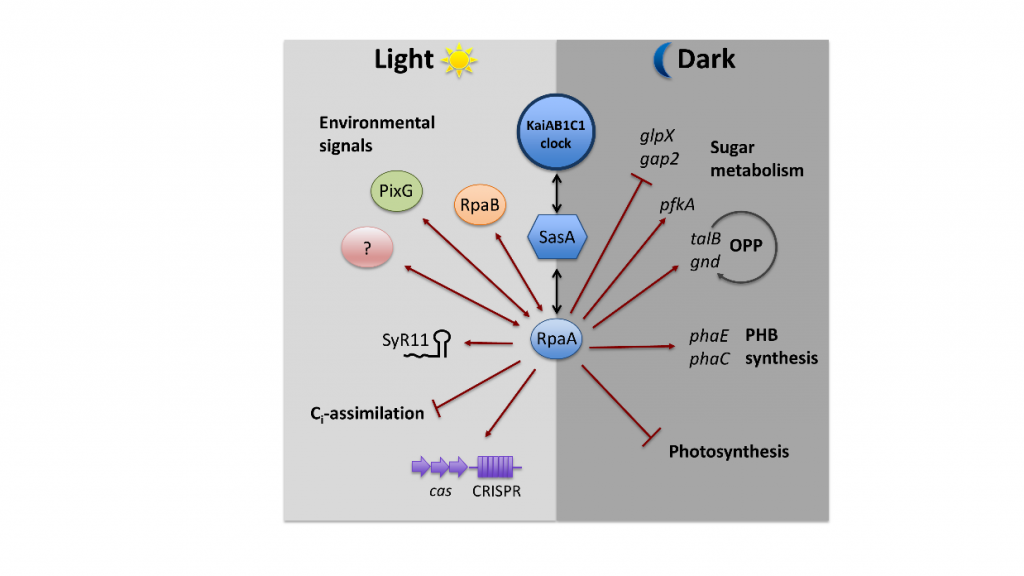
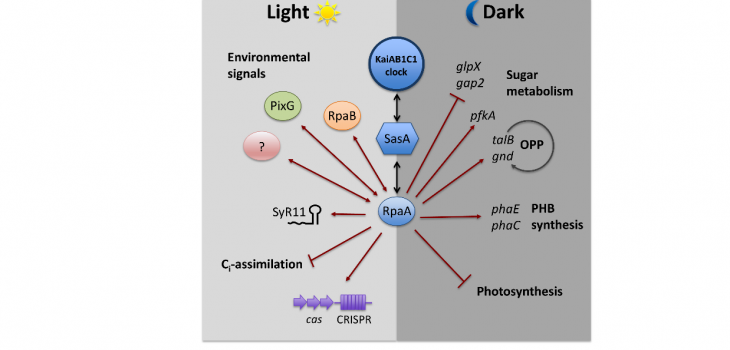
The output signaling of the KaiAB1C1 oscillator is mediated by a two-component system, consisting of the histidine kinase SasA and its response regulator RpaA. RpaA acts as a major transcription factor. The rpaA deletion mutant shows reduced viability in light-dark rhythms, especially under mixotrophic conditions and our results indicate an impaired carbon metabolism compared to the wild type.
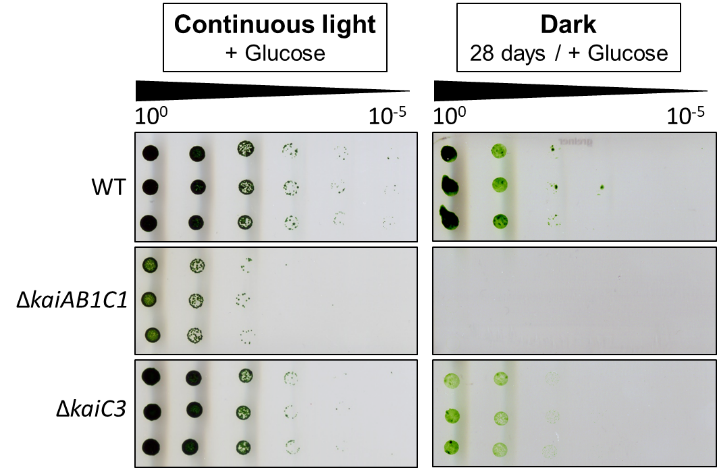
In this project we further aim at elucidating the function of the additional putative oscillators, which seem to utilize a RpaA-independent output pathway. The kaiC3 deletion mutant does not show a phenotype in light-dark rhythms, but in complete darkness during heterotrophic growth. In addition, in vitro and yeast two-hybrid data strongly imply cross talk between the distinct oscillators. However, function and direction of signaling remains unknown until now.
Funding:
- Joint DFG project together with Prof. Ilka Axmann: Non-standard bacterial clock systems
- Research unit FOR 2816: “SCyCode – The Autotrophy-Heterotrophy Switch in Cyanobacteria: Coherent Decision-Making at Multiple Regulatory Layers.”
Key publications:
Scheurer NM, Rajarathinam Y, Timm S, Köbler C, Kopka J, Hagemann M, Wilde A. (2021) Homologs of circadian clock proteins impact the metabolic switch between light and dark growth in the cyanobacterium Synechocystis sp. PCC 6803. Frontiers in Plant Science. 12, 1212.
Wiegard A, Köbler C, Oyama K, Dörrich AK, Azai C, Terauchi K, Wilde A, Axmann IM. (2020) Synechocystis KaiC3 displays temperature and KaiB dependent ATPase activity and is important for growth in darkness. J. Bacteriol. 202:e00478-19 doi: 10.1128/JB.00478-19. 31767776.
Köbler C, Schultz SJ, Kopp D, Voigt K, Wilde A. (2018) The role of the Synechocystis sp. PCC 6803 homolog of the circadian clock output regulator RpaA in day-night transitions. Mol. Microbiol. 110, 847-861.
Dörrich AK, Mitschke J, Siadat O, Wilde A. (2014) Deletion of the Synechocystis sp. PCC 6803 kaiAB1C1 gene cluster causes impaired cell growth under light-dark conditions. Microbiology 160, 2538-2550.
Axmann IM, Hertel S, Wiegard A, Dörrich AK, Wilde A. (2014) Diversity of KaiC-based timing systems in marine Cyanobacteria. Mar. Genomics. 14, 3-16.
Wiegard A, Dörrich AK, Deinzer HT, Beck C, Wilde A, Holtzendorff J, Axmann IM. (2013) Biochemical analysis of three putative KaiC clock proteins from Synechocystis sp. PCC 6803 suggests their functional divergence. Microbiology 159, 948-958.
Axmann, I.M., Dühring, U., Seeliger, L., Arnold, A., Vanselow, J.T., Kramer, A., Wilde, A. (2009) Biochemical evidence for a timing mechanism in Prochlorococcus. J. Bacteriol. 191, 5342-5347.